Research projects
General research interest
We are studying proteins and mechanisms that facilitate sculpting of cellular membranes into small vesicular structures, a necessity for trafficking and balancing of the membrane integrity but also for the replication of flaviviruses. Alterations and malfunctioning are implicated in a broad range of human diseases such as cancer, infection, obesity and lipid and muscle dystrophies.
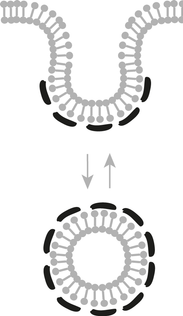
In particular, we are interested in the generation and stabilisation of atypical vesicles that remain associated with the donor membrane creating cavities in the membrane with extreme Gaussion curvature. Example of such cellular structures caveolae and flavivirus replication comparments.
Specific projects
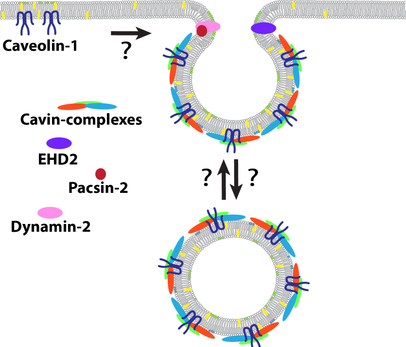
Caveolae biogenesis and dynamics
Caveolae are small relatively stable invaginations of the cell surface. They are found in most cells and are thought to play important roles in the control of lipid homeostasis, membrane tension and signalling. Caveolae dysfunction is strongly associated with tumourgenesis, cardiovascular disease, lipodystrophy and muscular dystrophy.
We are working on:
-Characterisation of the protein complexes and lipids that controls the stability of caveolae
-Architecture and assembly of the caveolae coat
-Influence of the lipid composition on the dynamics of caveolae
Role of membrane contact sites in lipid metabolism and fat cell expansion
Caveolae are highly abundant in tissues such as fat, muscle and vessels that handle large amounts of fatty acids. Clinical and experimental data show that caveolae heavily influence uptake and storage of fatty acids in cells. Caveolae are highly abundant in tissues that handle large amounts of fatty acids. However, the mechanisms by which caveolae act to control lipid sorting and trafficking is still not understood.
We are working on:
-Mechanism for how caveolae aid in the traffic of fatty acids
-Visualisation of contact sites between caveolae and the endoplasmic reticulum
Lipid metabolism and Breast cancer
Metabolic transformation is a hallmark of cancer cells which is key to sustain biomass production and proliferation. Triple negative breast cancer (TNBC) cells display such transformation through an upregulated fatty acid (FA) metabolism related to uptake of exogenous fatty acids. Yet, little is known about the precise metabolic derivatives and the enzymatic processes that drive this process.
We are working on:
-Determining the uptake rates and specific metabolic derivatives of FA in TNBC cells.
-characterisation of the mechanism and molecular machinery involved.
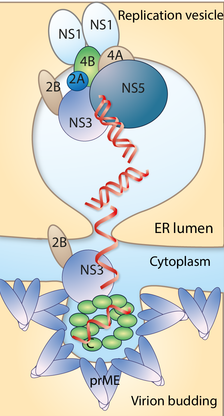
Biogenesis of replication vesicles by Tick-borne encephalitis virus
Tick-borne encephalitis virus belongs to the family of Flaviviruses, which cause sever disease worldwide. Directly upon viral entry the viral RNA genome is translated into a single polyprotein. The polyprotein is cleaved post-translation to generate three structural proteins (part of the virus coat) and seven nonstructural (NS) proteins (not part of the virus coat). The NS proteins are involved in protein cleavage, virus replication and the formation of virus factories although their exact functions are still unclear. A crucial part of the viral factory is the formation of small distinct invaginations of ~100 nm in diameter at the endoplasmatic reticulum. These so called replication vesicles, are induced by the viral proteins. Such cavities are open to the cytosol and virus RNA can be efficiently replicated here, well hidden away from the cellular response machinery specialized in detecting and degrading viral RNA. However, there is a lack of knowledge how viral proteins sculpt the membrane into replication vesicles.
We are working on:
-The structural mechanism for how the NS1 protein assemble on the lipid membrane
-The role of cellular host proteins in generation of replication compartments
Membrane sculpting during generation of new virus particles
Following replication, the RNA, the genome is transfered to a membrane budding sites that will generate a new virion. In this process, multiple copies of the capsid (C) protein recruit the newly synthesized viral genome to the membrane and form the nucleocapsid (NC). The NC buds into the ER lumen acquiring a host-derived lipid envelope with embedded surface proteins, the envelope (E) and pre-membrane (prM) proteins
We are working on:
-The structural mechanism for how the C protein interacts with the lipid membrane and RNA